New Features in RNA 3D Hub (August 2012)
RNA 3D Hub has been updated with new functionality making it easier to explore the RNA 3D Motif Atlas and the Loop Atlas.
RNA 3D Motif Atlas Updates
New features of the motif pages (for example, the sarcin-ricin motif):
- Similar motifs tab and a new motif comparison tool
Similar motifs are determined by computing minimum linkage between motif groups and shown on the new tab of the motif pages. Motif comparison tool can be launched in a new window by clicking the “Compare” link (example).
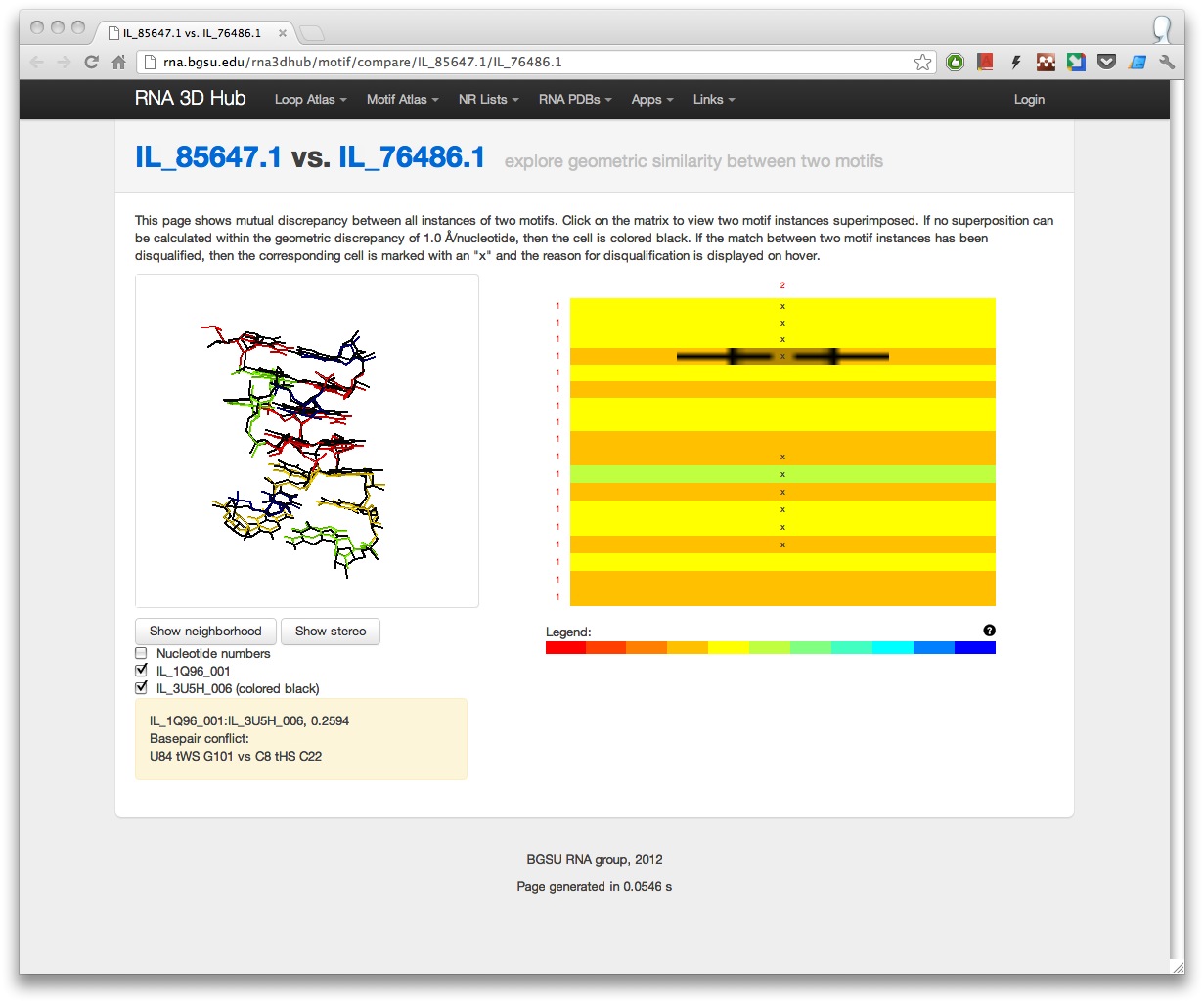
The comparison tool shows an interactive heatmap with geometric discrepancies between all instances of the two motifs. When the matrix is clicked, the Jmol window displays the superposition of the corresponding motif instances. If there is a structural conflict preventing some instances from being in the same motif group, then the cells in the heatmap are marked with an “x” and an explanation is provided on hover.
2. History tab
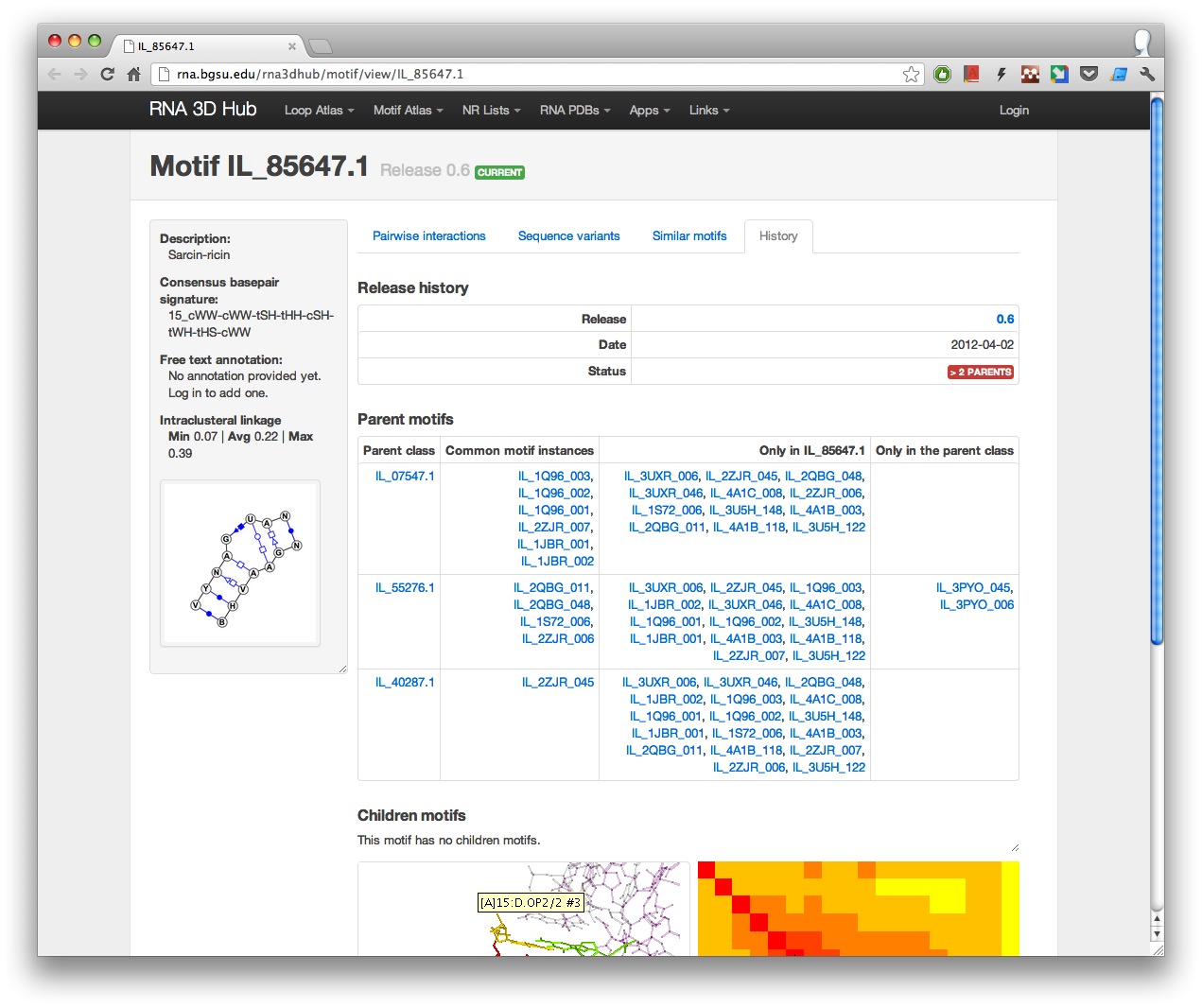
All releases in which the motif is present are listed on the page along with the parent and children motif classes, where applicable.
3. Links to individual loop instance pages. Now all motif pages link to the updated pages for each motif instance.
RNA Loop Atlas Updates
The RNA 3D Hub Loop Atlas contains all internal, hairpin or 3-way junction loops found in all RNA-containing 3D structures as of February 2012 (in the future the Loop Atlas will be updated regularly). Each loop has its own webpage. For example, a sarcin-ricin loop IL_1JBR_002 has the following stable url: http://rna.bgsu.edu/rna3dhub/loops/view/IL_1JBR_002.
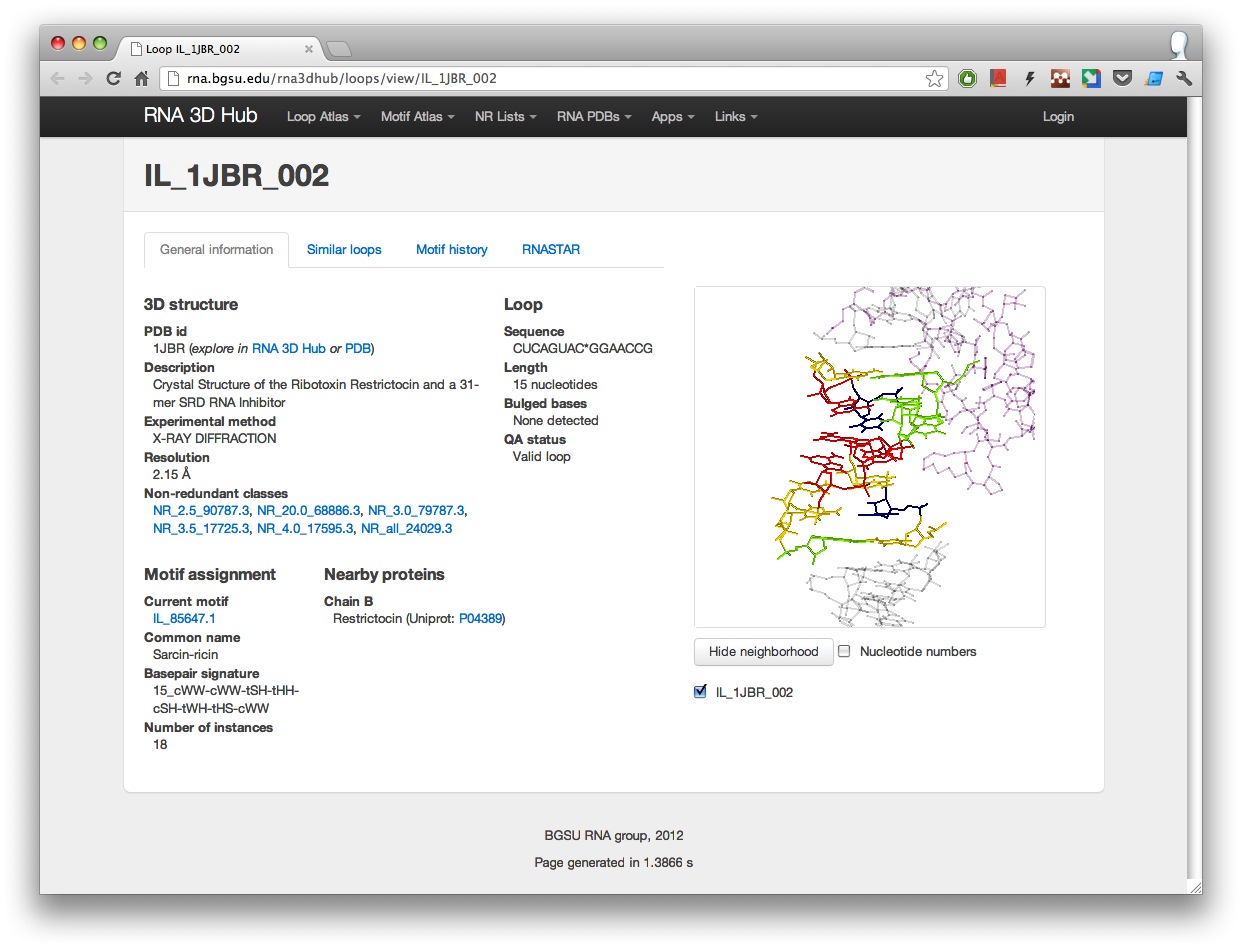
Now these pages include several new features:
- General overview of the PDB structure the loop is found in.
- Information about loop sequence and structure including bulged out bases.
- Current motif assignment where available.
- List of nearby proteins found in a 16A neighborhood. To view the proteins in 3D, click “Show neighborhood”. Proteins are colored pink.
- Similar loops tab, which interactively shows 3D loop superpositions with all similar loops within the geometric discrepancy of 1 A/nucleotide.
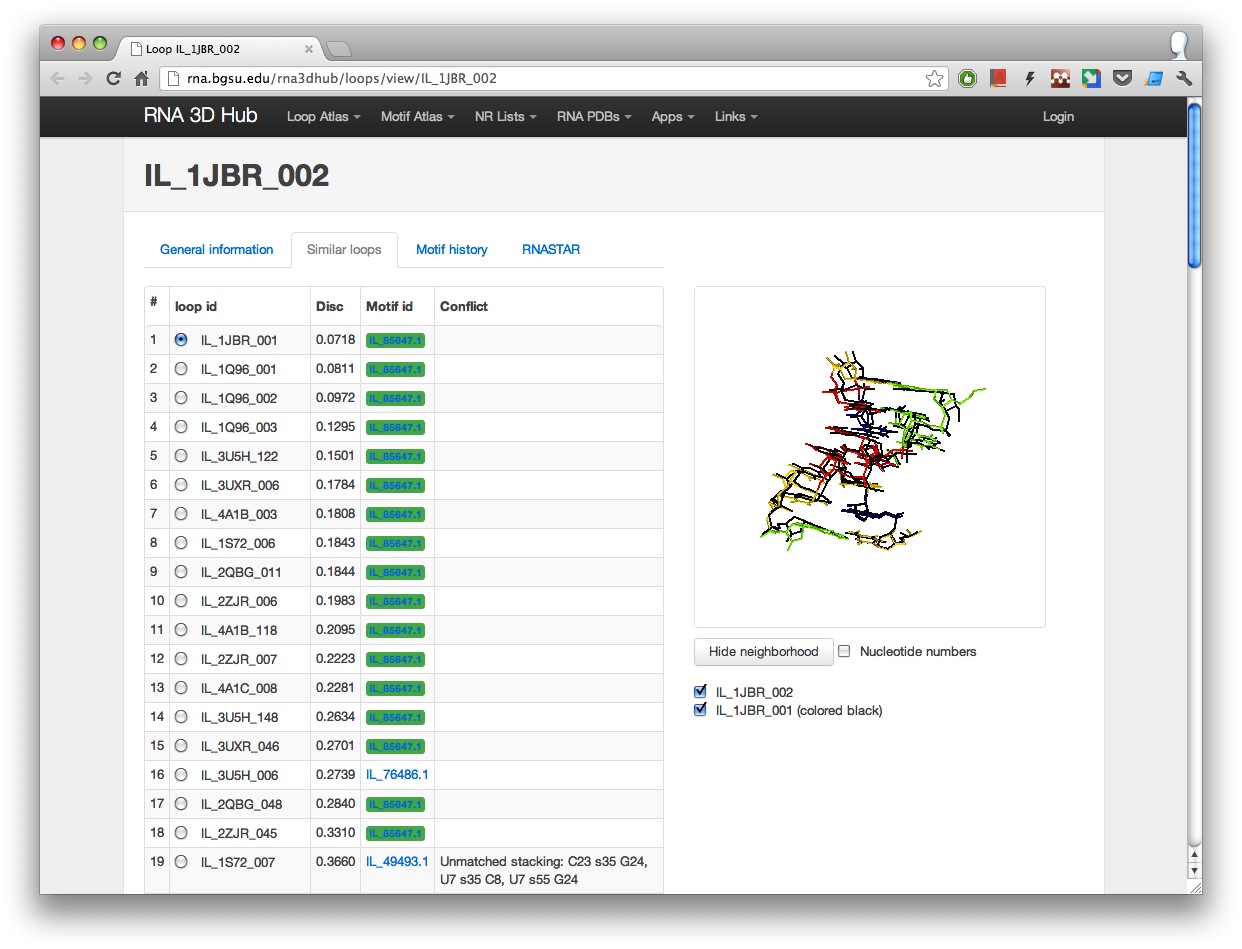
For each similar loop its motif assignment is also shown (loops belonging to the same motif as the main loop are highlighted in green). If two loops have conflicting structural features, a note explaining the reason behind this conflict is displayed. At this time similar loop comparison is limited to loops that have been assigned to RNA 3D motifs.
Updated: 12/02/2017 02:24AM
